Popcorn Hacks
Popcorn Hack 1: Find Students with Scores in a Range
pip install pandas
Collecting pandas
Using cached pandas-2.2.3-cp313-cp313-macosx_10_13_x86_64.whl.metadata (89 kB)
Requirement already satisfied: numpy>=1.26.0 in /Users/maryamabdul-aziz/vscode/maryam_2025/venv/lib/python3.13/site-packages (from pandas) (2.2.4)
Requirement already satisfied: python-dateutil>=2.8.2 in /Users/maryamabdul-aziz/vscode/maryam_2025/venv/lib/python3.13/site-packages (from pandas) (2.9.0.post0)
Requirement already satisfied: pytz>=2020.1 in /Users/maryamabdul-aziz/vscode/maryam_2025/venv/lib/python3.13/site-packages (from pandas) (2025.1)
Requirement already satisfied: tzdata>=2022.7 in /Users/maryamabdul-aziz/vscode/maryam_2025/venv/lib/python3.13/site-packages (from pandas) (2025.1)
Requirement already satisfied: six>=1.5 in /Users/maryamabdul-aziz/vscode/maryam_2025/venv/lib/python3.13/site-packages (from python-dateutil>=2.8.2->pandas) (1.17.0)
Using cached pandas-2.2.3-cp313-cp313-macosx_10_13_x86_64.whl (12.5 MB)
Installing collected packages: pandas
Successfully installed pandas-2.2.3
[1m[[0m[34;49mnotice[0m[1;39;49m][0m[39;49m A new release of pip is available: [0m[31;49m25.0[0m[39;49m -> [0m[32;49m25.0.1[0m
[1m[[0m[34;49mnotice[0m[1;39;49m][0m[39;49m To update, run: [0m[32;49mpip install --upgrade pip[0m
Note: you may need to restart the kernel to use updated packages.
import pandas as pd
student_data = pd.DataFrame({
'Name': ['Aditi', 'Brady', 'Charlotte', 'Daniel', 'Emily'],
'Score': [92, 84, 78, 95, 82],
'Grade': ['A', 'B', 'C', 'A', 'B']
})
# Complete the function to find all students with scores between min_score and max_score
def find_students_in_range(df, min_score, max_score):
return df[(df['Score'] >= min_score) & (df['Score'] <= max_score)]
print(find_students_in_range(student_data, 80, 90))
Name Score Grade
1 Brady 84 B
4 Emily 82 B
PopCorn Hack 2: Calculate Letter Grades
import pandas as pd
student_data = pd.DataFrame({
'Name': ['Aditi', 'Brady', 'Charlotte', 'Daniel', 'Emily'],
'Score': [92, 84, 78, 95, 82],
'Grade': ['A', 'B', 'C', 'A', 'B']
})
# Complete the function to add a 'Letter' column based on numerical scores
def add_letter_grades(df):
def get_letter(score):
if score >= 90:
return 'A'
elif score >= 80:
return 'B'
elif score >= 70:
return 'C'
elif score >= 60:
return 'D'
else:
return 'F'
df['Letter'] = df['Score'].apply(get_letter)
return df
add_letter_grades(student_data)
| Name | Score | Grade | Letter | |
|---|---|---|---|---|
| 0 | Aditi | 92 | A | A |
| 1 | Brady | 84 | B | B |
| 2 | Charlotte | 78 | C | C |
| 3 | Daniel | 95 | A | A |
| 4 | Emily | 82 | B | B |
PopCorn Hack 3: Find the Mode in a Series
# Complete the function to find the most common value in a series
def find_mode(series):
mode = series.mode()
return mode
find_mode(pd.Series([1, 2, 2, 3, 4, 2, 5]))
0 2
dtype: int64
Homework Hacks
For this homework, you’ll work with a dataset of student performance and implement various list algorithms using both Python lists and Pandas.
Dataset: Pima Indians Diabetes Info
Algorithms
import pandas as pd
datas = pd.read_csv('/Users/maryamabdul-aziz/vscode/maryam_2025/_notebooks/data/diabetes.csv')
# Highest and lowest BMI
def get_extreme_bmi(datas):
highest_bmi = datas[datas['BMI'] == datas['BMI'].max()]
lowest_bmi = datas[datas['BMI'] == datas['BMI'].min()]
return highest_bmi, lowest_bmi
# Difference between max and min for Glucose, BloodPressure, and BMI
def add_max_min_diff(datas):
datas['MaxMinDiff'] = datas[['Glucose', 'BloodPressure', 'BMI']].max(axis=1) - \
datas[['Glucose', 'BloodPressure', 'BMI']].min(axis=1)
return datas
# Patients with Glucose above the average
def get_above_avg_glucose(datas):
avg_glucose = datas['Glucose'].mean()
return datas[datas['Glucose'] > avg_glucose]
# Group by AgeGroup and Outcome, then calculate mean BMI and Glucose
def group_by_age_outcome(datas):
datas = datas.copy()
datas['AgeGroup'] = pd.cut(datas['Age'], bins=[20, 30, 40, 50, 60, 100],
labels=['20s', '30s', '40s', '50s', '60+'], right=False)
grouped = datas.groupby(['AgeGroup', 'Outcome'])[['BMI', 'Glucose']].mean().reset_index()
return grouped
# Correlation between Age and BMI
def age_bmi_correlation(datas):
return datas['Age'].corr(datas['BMI'])
# Age group with highest average glucose
def age_group_with_highest_glucose(datas):
datas = datas.copy()
datas['AgeGroup'] = pd.cut(datas['Age'], bins=[20, 30, 40, 50, 60, 100],
labels=['20s', '30s', '40s', '50s', '60+'], right=False)
glucose_by_age = datas.groupby('AgeGroup')['Glucose'].mean()
return glucose_by_age.idxmax(), glucose_by_age
# Percentage of individuals with BMI > 30 (obese)
def percent_obese(datas):
obese_count = datas[datas['BMI'] > 30].shape[0]
total_count = datas.shape[0]
return (obese_count / total_count) * 100
highest, lowest = get_extreme_bmi(datas)
datas = add_max_min_diff(datas)
glucose_above_avg = get_above_avg_glucose(datas)
grouped_stats = group_by_age_outcome(datas)
correlation = age_bmi_correlation(datas)
top_age_group, glucose_age_distribution = age_group_with_highest_glucose(datas)
obese_pct = percent_obese(datas)
print(highest, lowest)
print(datas)
print(glucose_above_avg)
print(grouped_stats)
print(correlation)
print(top_age_group, glucose_age_distribution)
print(obese_pct)
Pregnancies Glucose BloodPressure SkinThickness Insulin BMI \
177 0 129 110 46 130 67.1
DiabetesPedigreeFunction Age Outcome
177 0.319 26 1 Pregnancies Glucose BloodPressure SkinThickness Insulin BMI \
9 8 125 96 0 0 0.0
49 7 105 0 0 0 0.0
60 2 84 0 0 0 0.0
81 2 74 0 0 0 0.0
145 0 102 75 23 0 0.0
371 0 118 64 23 89 0.0
426 0 94 0 0 0 0.0
494 3 80 0 0 0 0.0
522 6 114 0 0 0 0.0
684 5 136 82 0 0 0.0
706 10 115 0 0 0 0.0
DiabetesPedigreeFunction Age Outcome
9 0.232 54 1
49 0.305 24 0
60 0.304 21 0
81 0.102 22 0
145 0.572 21 0
371 1.731 21 0
426 0.256 25 0
494 0.174 22 0
522 0.189 26 0
684 0.640 69 0
706 0.261 30 1
Pregnancies Glucose BloodPressure SkinThickness Insulin BMI \
0 6 148 72 35 0 33.6
1 1 85 66 29 0 26.6
2 8 183 64 0 0 23.3
3 1 89 66 23 94 28.1
4 0 137 40 35 168 43.1
.. ... ... ... ... ... ...
763 10 101 76 48 180 32.9
764 2 122 70 27 0 36.8
765 5 121 72 23 112 26.2
766 1 126 60 0 0 30.1
767 1 93 70 31 0 30.4
DiabetesPedigreeFunction Age Outcome MaxMinDiff
0 0.627 50 1 114.4
1 0.351 31 0 58.4
2 0.672 32 1 159.7
3 0.167 21 0 60.9
4 2.288 33 1 97.0
.. ... ... ... ...
763 0.171 63 0 68.1
764 0.340 27 0 85.2
765 0.245 30 0 94.8
766 0.349 47 1 95.9
767 0.315 23 0 62.6
[768 rows x 10 columns]
Pregnancies Glucose BloodPressure SkinThickness Insulin BMI \
0 6 148 72 35 0 33.6
2 8 183 64 0 0 23.3
4 0 137 40 35 168 43.1
8 2 197 70 45 543 30.5
9 8 125 96 0 0 0.0
.. ... ... ... ... ... ...
759 6 190 92 0 0 35.5
761 9 170 74 31 0 44.0
764 2 122 70 27 0 36.8
765 5 121 72 23 112 26.2
766 1 126 60 0 0 30.1
DiabetesPedigreeFunction Age Outcome MaxMinDiff
0 0.627 50 1 114.4
2 0.672 32 1 159.7
4 2.288 33 1 97.0
8 0.158 53 1 166.5
9 0.232 54 1 125.0
.. ... ... ... ...
759 0.278 66 1 154.5
761 0.403 43 1 126.0
764 0.340 27 0 85.2
765 0.245 30 0 94.8
766 0.349 47 1 95.9
[349 rows x 10 columns]
AgeGroup Outcome BMI Glucose
0 20s 0 29.852885 106.503205
1 20s 1 37.101190 140.642857
2 30s 0 30.923596 113.348315
3 30s 1 34.285526 139.315789
4 40s 0 33.318868 109.509434
5 40s 1 35.676923 136.984615
6 50s 0 30.221739 123.782609
7 50s 1 32.094118 151.441176
8 60+ 0 27.165217 131.391304
9 60+ 1 31.755556 155.777778
0.03624187009229404
50s AgeGroup
20s 113.744949
30s 125.309091
40s 124.644068
50s 140.280702
60+ 138.250000
Name: Glucose, dtype: float64
60.546875
/var/folders/93/y55vzx413xxdrd4m_6cv_rmm0000gn/T/ipykernel_13033/3696993698.py:26: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning.
grouped = datas.groupby(['AgeGroup', 'Outcome'])[['BMI', 'Glucose']].mean().reset_index()
/var/folders/93/y55vzx413xxdrd4m_6cv_rmm0000gn/T/ipykernel_13033/3696993698.py:38: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning.
glucose_by_age = datas.groupby('AgeGroup')['Glucose'].mean()
Analytical Questions
import pandas as pd
datas = pd.read_csv('/Users/maryamabdul-aziz/vscode/maryam_2025/_notebooks/data/diabetes.csv')
def master_function(datas):
# What is the correlation between diabetes pedigree and other factors?
preg = datas['DiabetesPedigreeFunction'].corr(datas['Pregnancies'])
glucose = datas['DiabetesPedigreeFunction'].corr(datas['Glucose'])
bp = datas['DiabetesPedigreeFunction'].corr(datas['BloodPressure'])
st = datas['DiabetesPedigreeFunction'].corr(datas['SkinThickness'])
ins = datas['DiabetesPedigreeFunction'].corr(datas['Insulin'])
bmi = datas['DiabetesPedigreeFunction'].corr(datas['BMI'])
age = datas['DiabetesPedigreeFunction'].corr(datas['Age'])
print(preg, glucose, bp, st, ins, bmi, age)
# Which Age group has the highest average Glucose?
# Create AgeGroup column
datas = datas.copy()
datas['AgeGroup'] = pd.cut(
datas['Age'],
bins=[20, 30, 40, 50, 60, 100],
labels=['20s', '30s', '40s', '50s', '60+'],
right=False
)
glucose_by_age = datas.groupby('AgeGroup')['Glucose'].mean()
highest_glucose_group = glucose_by_age.idxmax()
print(highest_glucose_group)
# What percentage of people have BMI above 30 (obese)?
obese = datas[datas['BMI'] > 30]
obese_percentage = len(obese) / len(datas) * 100
print(obese_percentage)
master_function(datas)
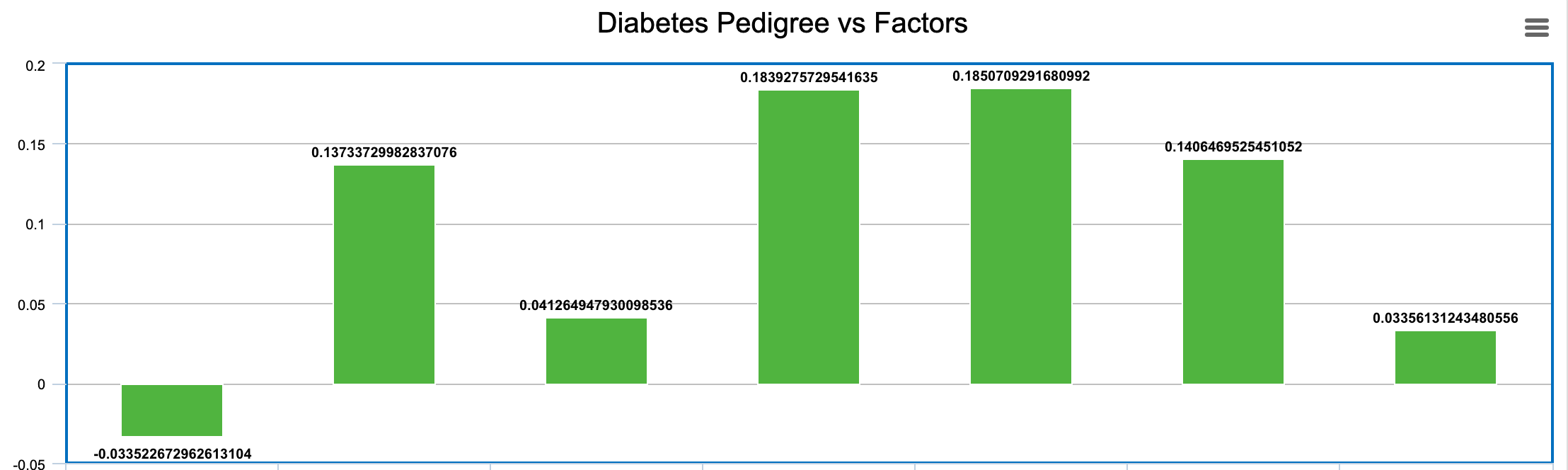
-0.033522672962613104 0.13733729982837076 0.041264947930098536 0.1839275729541635 0.1850709291680992 0.1406469525451052 0.03356131243480556
50s
60.546875
/var/folders/93/y55vzx413xxdrd4m_6cv_rmm0000gn/T/ipykernel_13033/2651657759.py:26: FutureWarning: The default of observed=False is deprecated and will be changed to True in a future version of pandas. Pass observed=False to retain current behavior or observed=True to adopt the future default and silence this warning.
glucose_by_age = datas.groupby('AgeGroup')['Glucose'].mean()
Chart:
SQL
import sqlite3
import pandas as pd
datas = pd.read_csv('/Users/maryamabdul-aziz/vscode/maryam_2025/_notebooks/data/diabetes.csv')
conn = sqlite3.connect(":memory:")
datas.to_sql("diabetes", conn, index=False, if_exists="replace")
query = "SELECT Outcome, AVG(Glucose) AS avg_glucose FROM diabetes GROUP BY Outcome"
result = pd.read_sql_query(query, conn)
print(result)
import sqlite3
import pandas as pd
data = pd.read_csv('/Users/maryamabdul-aziz/vscode/maryam_2025/_notebooks/data/diabetes.csv')
conn = sqlite3.connect(':memory:')
data.to_sql('diabetes', conn, index=False, if_exists='replace')
query = "SELECT * FROM diabetes WHERE Outcome = 1"
diabetic_patients = pd.read_sql_query(query, conn)
print(diabetic_patients)
query = """
SELECT AVG(Age) as AvgAge, AVG(BMI) as AvgBMI, COUNT(*) as Count
FROM diabetes
WHERE Outcome = 1
"""
diabetic_stats = pd.read_sql_query(query, conn)
print(diabetic_stats)
query = """
SELECT Outcome, COUNT(*) as Count
FROM diabetes
GROUP BY Outcome
"""
outcome_distribution = pd.read_sql_query(query, conn)
print(outcome_distribution)
Pregnancies Glucose BloodPressure SkinThickness Insulin BMI \
0 6 148 72 35 0 33.6
1 8 183 64 0 0 23.3
2 0 137 40 35 168 43.1
3 3 78 50 32 88 31.0
4 2 197 70 45 543 30.5
.. ... ... ... ... ... ...
263 1 128 88 39 110 36.5
264 0 123 72 0 0 36.3
265 6 190 92 0 0 35.5
266 9 170 74 31 0 44.0
267 1 126 60 0 0 30.1
DiabetesPedigreeFunction Age Outcome
0 0.627 50 1
1 0.672 32 1
2 2.288 33 1
3 0.248 26 1
4 0.158 53 1
.. ... ... ...
263 1.057 37 1
264 0.258 52 1
265 0.278 66 1
266 0.403 43 1
267 0.349 47 1
[268 rows x 9 columns]
AvgAge AvgBMI Count
0 37.067164 35.142537 268
Outcome Count
0 0 500
1 1 268
- Discuss the advantages and disadvantages of using SQL versus Pandas for data analysis, focusing on performance, readability, and ease of use.
SQL is better for large datasets that need complicated queries and aggregations. It’s better for structure but can become difficult to use for advanced data manipulation. Pandas is better for smaller datasets because it’s flexible, allowing for quick analysis with Python. However, it can struggle with large data and become less efficient for complicated queries.